COMPETENT HOST
● Since DNA is a `color{Violet}"hydrophilic molecule"`, it cannot pass through `color{Violet}"cell membranes"`.
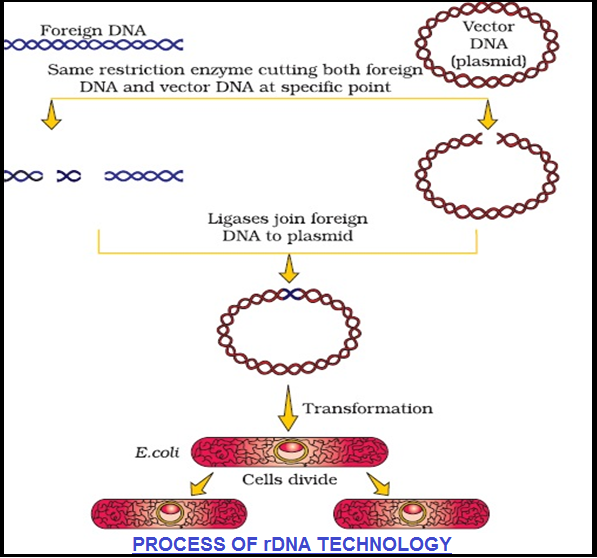
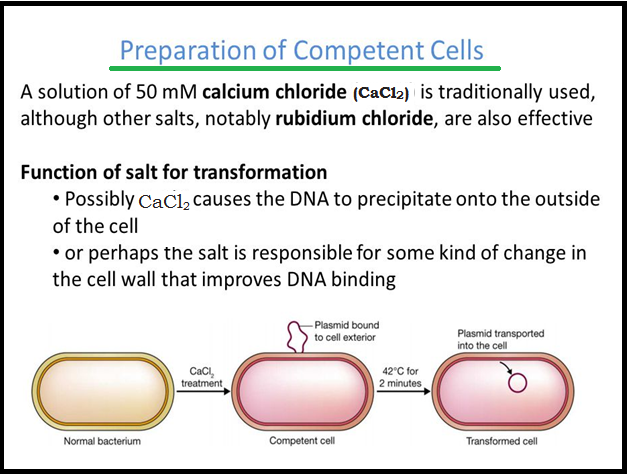
● In order to `color{Violet}"force bacteria"` to take up the plasmid, the bacterial cells must first be `color{Brown}"made ‘competent"` to take up DNA.
● This is done by treating them with a specific concentration of a `color{Violet}"divalent cation"`, such as `color{Violet}"calcium"`, which increases the efficiency with which DNA enters the bacterium through `color{Violet}"pores in its cell wall"`.
● Recombinant DNA can then be forced into such cells by `color{Violet}"incubating the cells"` with recombinant DNA on `color{Violet}"ice"`, followed by placing them briefly at `color{Violet}"42° C (heat shock)"`, and then putting them back on ice.
● This `color{Violet}"enables the bacteria"` to take up the `color{Violet}"recombinant DNA"`.
● This is not the only way to `color{Violet}"introduce alien DNA"` into host cells.
● In a method known as `color{Brown}"micro-injection"`, recombinant DNA is `color{Violet}"directly injected"` into the nucleus of an animal cell.

● In another method, `color{Violet}"suitable for plants"`, cells are bombarded with `color{Violet}"high velocity micro-particles"` of `color{Violet}"gold or tungsten"` coated with DNA in a method known as `color{Brown}"biolistics or gene gun"`.

● And the last method uses `color{Violet}"disarmed pathogen"` vectors, which when allowed to infect the cell, transfer the recombinant DNA into the host.
● In order to `color{Violet}"force bacteria"` to take up the plasmid, the bacterial cells must first be `color{Brown}"made ‘competent"` to take up DNA.
● This is done by treating them with a specific concentration of a `color{Violet}"divalent cation"`, such as `color{Violet}"calcium"`, which increases the efficiency with which DNA enters the bacterium through `color{Violet}"pores in its cell wall"`.
● Recombinant DNA can then be forced into such cells by `color{Violet}"incubating the cells"` with recombinant DNA on `color{Violet}"ice"`, followed by placing them briefly at `color{Violet}"42° C (heat shock)"`, and then putting them back on ice.
● This `color{Violet}"enables the bacteria"` to take up the `color{Violet}"recombinant DNA"`.
● This is not the only way to `color{Violet}"introduce alien DNA"` into host cells.
● In a method known as `color{Brown}"micro-injection"`, recombinant DNA is `color{Violet}"directly injected"` into the nucleus of an animal cell.

● In another method, `color{Violet}"suitable for plants"`, cells are bombarded with `color{Violet}"high velocity micro-particles"` of `color{Violet}"gold or tungsten"` coated with DNA in a method known as `color{Brown}"biolistics or gene gun"`.

● And the last method uses `color{Violet}"disarmed pathogen"` vectors, which when allowed to infect the cell, transfer the recombinant DNA into the host.